In this tutorial we consider a phantom consisting of conductive inclusions in the unit ball with a known background conductivity.
| Inclusion | Center | Radii | Axes | Conductivity |
|---|---|---|---|---|
| Ball | (-0.09,-0.55,0) | r = 0.273 | 2 | |
| Left spheroid | 0.55(-sin(5π/12),cos(5π/12),0) | r1 = 0.468 r2 = 0.234 r3 = 0.234 |
(cos(5π/12),sin(5π/12),0) (-sin(5π/12),cos(5π/12),0) (0,0,1) |
0.5 |
| Right spheroid | 0.45(sin(5π/12),cos(5π/12),0) | r1 = 0.546 r2 = 0.273 r3 = 0.273 |
(cos(5π/12),-sin(5π/12),0) (sin(5π/12),cos(5π/12),0) (0,0,1) |
0.5 |
Table of Contents
Forward problem solver
Assuming crEITive has been installed successfully and initialized by run startup we start by defining the needed constants:
% Orientation rotang=5*pi/12; % Prolate spheroid on the right % Center xr0=sin(rotang)*0.45; yr0=cos(rotang)*0.45; zr0=0; % Radius rr=0.42; % Conductivity cr=0.5; % Prolate spheroid on the left xl0=-sin(rotang)*0.55; yl0=cos(rotang)*0.55; zl0=0; % Smallest radius rl=0.36; % Conductivity cl=0.5; % Axes A = [cos(rotang),sin(rotang),0;-sin(rotang),cos(rotang),0;0,0,1]; % Ball % Center xb0=-0.09; yb0=-0.55; zb0=0; % Radius rb=0.21; % Conductivity cb=2;
Creating the conductivity objects is then straightforward. Currently crEITive supports two classes of conductivity: piecewise constant and radial. We have to choose the number of quadrature points for each of the inclusions.
% Possible Classes:
% Piecewise constant:
% - PcBallConductivity ( center , radius , amplitude , n )
% - PcEllipsoidConductivity ( center , radii , axes , amplitude , n )
% Radial:
% - RadialBallConductivity ( radius , amplitude , innerextent )
% Quadrature points: 2(n+1)² gives the number of quadrature points on
% the unit sphere. The quadrature integrates exactly spherical harmonics
% of degree less than or equal to n. These points are mapped onto the
% ellipsoid boundary.
n = 10;
% Create different conductivity elements for pcc
SRpc = PcEllipsoidConductivity([xr0,yr0,zr0], ...
[1.3*rr, 0.65*rr, 0.65*rr], A', cr, n);
SLpc = PcEllipsoidConductivity([xl0,yl0,zl0], ...
[1.3*rl, 0.65*rl, 0.65*rl], A, cl, n);
B1pc = PcBallConductivity([xb0,yb0,zb0], 1.3*rb, cb, n);
% Collect total conductivity data
conddata_pc = MakeConductivityData(SRpc,SLpc,B1pc);
% Plot
PlotPCPhantom(conddata_pc)
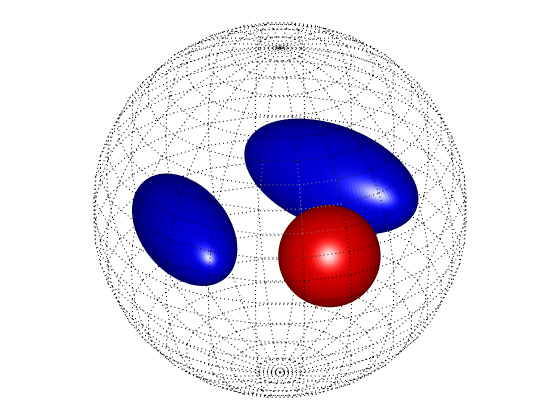
To compute the Dirichlet-to-Neumann map of this conductivity distribution we use a function implementing a boundary element method, see Research. For this example we choose the following parameters:
%% Forward map parameters
% Maximal degree of spherical harmonics in the
% computation of the Dirichlet-to-Neumann map.
nd = 10;
% Mesh used to save the conductivity and q at points.
% Useful to compare the conductivity and reconstruction
% at points of a gmsh mesh.
mesh = 'ball_0p1_3D.msh'; % choose mesh from crEITive/mesh/ folder
savecond = 1; % {0,1} - save conductivity data or not?
saveq = 1; % {0,1} - save q data or not?
parallel = 1; % {0,1} - parallelize or not? (Requires openMP)
% (optional) Define your own script for running on a cluster
clusterscript = 0;
% Unique id number for your forward computation
dnmapid = 1;
The following command then saves all the provided information into a command file in crEITive/commands/dnmap_commands.
% complex - {0,1} whether phantom is complex or not
% commands - path and name of command file
% log - path and name of log file
[complex, commands, log] = WritePcCommandsFile(conddata_pc,nd,dnmapid,...
savecond,saveq,mesh);
You may either simply run ./bin/pcc [commands] [log] from your terminal window in the folder crEITive. Or you can provide a script that submits the command to a computing cluster. See submitToDTUCluster.m for an example; in this case you also provide an email for updates.
% Start StartDNMAP(method, parallel, complex, commands, log, clusterscript, email)
When the program is done, the resulting Dirichlet-to-Neumann map is saved in the folder crEITive/results/dnmap and is identifiable with the id number.
Reconstruction
Given a Dirichlet-to-Neumann map in the data type described under Data storage we may reconstruct a conductivity using a variety of CGO-based methods (see Research):
% Method used for the reconstruction % - calderon for a reconstruction with Calderón approximation. % - texp for a reconstruction with texp (Born) approximation. % - t0 for a reconstruction with t0 approximation. % - t for a reconstruction with full algorithm. reconmethod = 'texp';
There is a number of reconstruction parameters pertaining to the so-called scattering transform and the regularization of the inverse map
% Computation of inverse Fourier transform
% - ifft for an Inverse Fast Fourier Transform
ift = 'ifft';
% Resolution of grid where the scattering transform is computed
ngrid = 11;
% Truncation radius (maximum frequency of scattering transform)
% We must have truncrad <= pi*(ngrid-1)^2/(2*ngrid) by Shannon Sampling
truncrad = 9;
% Complex frequency (zeta) size and type.
% - |zeta| = pkappa*truncrad/sqrt(2) if fixed = 1.
% - |zeta| = pkappa*|xi|/sqrt(2) for each xi if fixed = 0.
fixed = 1; % {0,1} choose zeta fixed (1) or proportional (0)
pkappa = 1; % pkappa >= 1
Similar to above we define additional variables and give our reconstruction a unique id.
% Compute conductivity and q on mesh
mesh = 'ball_0p05_3D.msh'; % choose mesh from crEITive/mesh/ folder
dnmapid = 1; % unique dnmap integer id number of dnmap data
reconid = 1; % unique reconstruction integer id number
parallel = 1; % {0,1} - parallelize or not?
cluster = 0;
Computing the inverse map is then straightforward and completely analogous to the forward map
% Commands
[complex, commands, log] = WriteCommandsFile(reconmethod,ift,ngrid,...
truncrad,fixed,pkappa,mesh,reconid,dnmapid);
% Start
StartEIT(parallel, complex, commands, log, clusterscript, email);
A number of results is then saved in crEITive/results: conductivity, potential q, scattering transform qhat and trace of CGO solutions, all identified by the reconstruction id. We can make a second reconstruction with the full algorithm:
% Full algorithm
reconmethod = 't';
reconid = 2;
% Commands
[complex, commands, log] = WriteCommandsFile(reconmethod,ift,ngrid,...
truncrad,fixed,pkappa,mesh,reconid,dnmapid);
% Start
StartEIT(parallel, complex, commands, log, clusterscript, email);
Note the results are named with a specific string (conductivity_2_t_nd_10_fixed_ifft_ngrid_11_kap_1_dnmap_1.mat) containing information on the reconstruction parameters.
Plots
The results of the reconstruction function allows for simple plots. In this case we compare the reconstructions above corresponding to reconid = 1 and reconid = 2. To make the plots appealing we restrict the plot function to comparing an even amount of results. With Plot2D we may view a 2D slice of our three-dimensional conductivity reconstruction.
% You may choose from {'conductivity','q','qhat'}
type = 'conductivity';
reconid = [1,2]; % reconstruction id
dnmapid = 0; % include true data by giving dnmapid
v = [0,0,1]; % normal vector to slice
h = 0.01; % stepsize
span = [0.65,1.7]; % span of type
[ha, pos] = Plot2D(type, reconid, dnmapid, v, h, span);
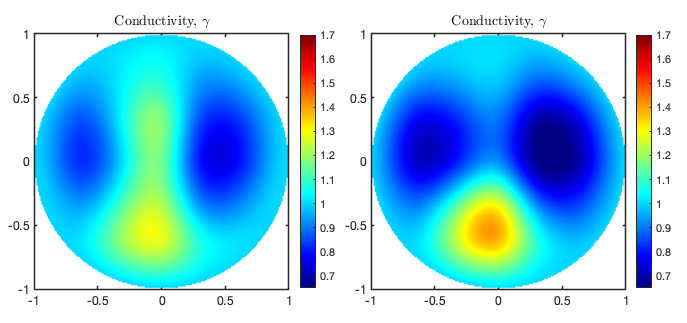
The plot functions returns a figure handle, allowing for simple modification of the figure. Similarly we may want to plot one-dimensional restrictions of results along straight lines. Using Plot1D we can compare several restrictions in subplots.
% You may choose from {'conductivity','q','qhat'}
type = 'conductivity';
% comp_reconid is a matrix with columns representing
% plots, in which the elements are ids to types we
% want to compare in 1D plot
comp_reconid = cat(2,[1;2],[1;2]);
% include true data by giving dnmapids in a vector
include_true = [1,1];
% comp_what is a cell list length == size(comp_reconid,2)
% of "comparison"-strings. One "comparison"-string in
% {'method','ift','ngrid','fixed','pkappa'} for each
% column in comp_reconid.
comp_what = {'reconmethod','reconmethod'};
The columns of comp_reconid corresponds to different subplots and the elements in those vectors consists of the reconstructions we wish to compare. In this example we consider two identical subplots comparing reconstructions reconid = 1 and reconid = 2 with the true phantom.
% Restriction of the conductivity to a line requires a
% direction v and a point center
v = [1,0,0];
center = [0,0,0];
h = 0.01; % stepsize
span = [0.4,1.3]; % span of type
[ha, pos] = Plot1D(type,comp_reconid,include_true, ...
comp_what,v,center,h,span);
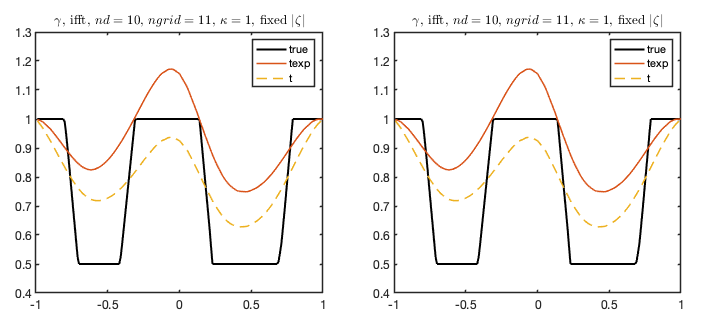
We may also plot several two-dimensional slices of a conductivity distribution in the same plot.
% 3D plots supports only conductivity type of data
reconid = 1; % reconstruction id
dnmapid = 0; % include true data by giving dnmapid
% Slices are given as for the Matlab slice function
xslice = [-0.6,-0.05,0.6]; % slice vector x
yslice = []; % slice vector y
zslice = 0; % slice vector z
h = 0.01; % stepsize
span = [0.8,1.4]; % span of type
[ha, pos] = Plot3Dc(reconid, dnmapid, xslice, yslice, ...
zslice, h, span);
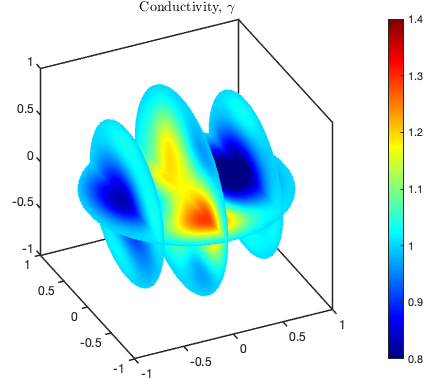
Data storage
This document describes how the data are stored in the different results files.
Additional functions
The crEITive software tool includes a number of auxiliary Matlab functions for plotting, reading, writing and computing data:
- plot_piecewise_constant_heart_lungs.m – plots a 3D view of the piecewise constant hearts-lungs phantom used here
- tight_subplot.m – for tight subplots by Pekka Kumpulainen
- read_conductivity.m – reads conductivity.mat files
- read_gmsh_mesh.m – reads gmsh mesh file
- read_q.m
- read_qhat.m
- read_dnmap.m
- snorm.m – computes approximation of L^2-based Sobolev H^s operator norm of matrix approximation of operator
- add_noise_dnmap.m – adds noise to Dirichlet-to-Neumann map according to s-norm
- write_dnmap.m – writes matrix to Dirichlet-to-Neumann map file according to data storage
- gauss_legendre.m – computes nodes and weights for Gauss-Legendre quadrature rule of order n on [-1,1]
- quad_points_unit_sphere.m – computes quadrature points on the unit sphere